Group 6 TMDs¶
Tight-binding models for group 6 transition metal dichalcogenides (TMD).
-
monolayer_3band(name, override_params=None)¶ Monolayer of a group 6 TMD using the nearest-neighbor 3-band model
Parameters: name : str
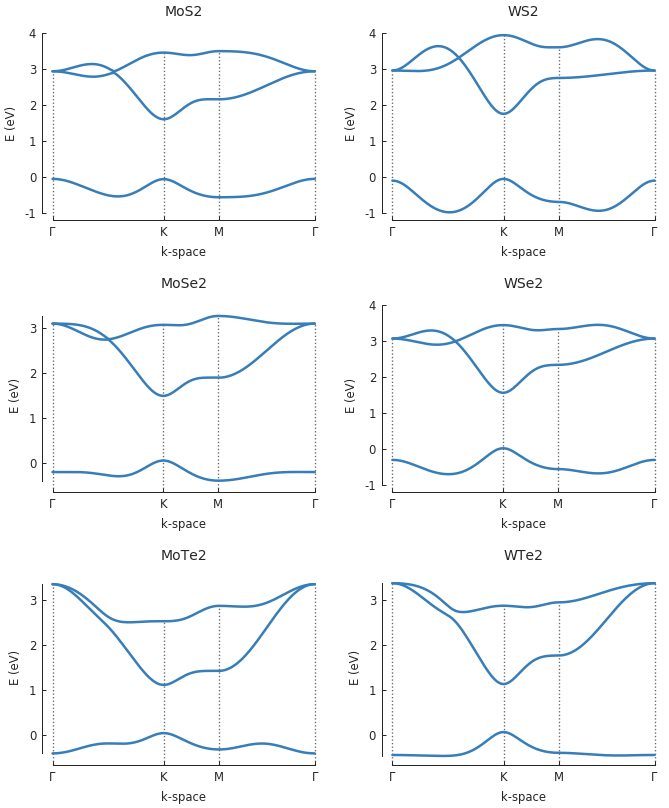
Name of the TMD to model. The available options are: MoS2, WS2, MoSe2, WSe2, MoTe2, WTe2. The relevant tight-binding parameters for these materials are given by https://doi.org/10.1103/PhysRevB.88.085433.
override_params : Optional[dict]
Replace or add new material parameters. The dictionary entries must be in the format
"name": [a, eps1, eps2, t0, t1, t2, t11, t12, t22].Examples
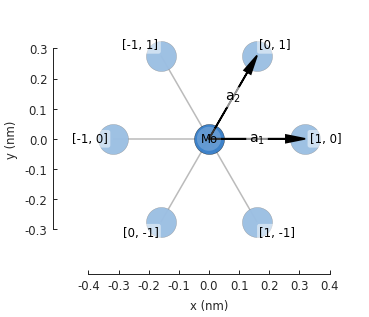
from pybinding.repository import group6_tmd group6_tmd.monolayer_3band("MoS2").plot()
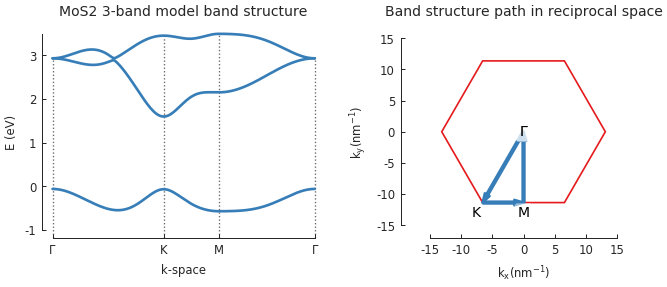
model = pb.Model(group6_tmd.monolayer_3band("MoS2"), pb.translational_symmetry()) solver = pb.solver.lapack(model) k_points = model.lattice.brillouin_zone() gamma = [0, 0] k = k_points[0] m = (k_points[0] + k_points[1]) / 2 plt.figure(figsize=(6.7, 2.3)) plt.subplot(121, title="MoS2 3-band model band structure") bands = solver.calc_bands(gamma, k, m, gamma) bands.plot(point_labels=[r"$\Gamma$", "K", "M", r"$\Gamma$"]) plt.subplot(122, title="Band structure path in reciprocal space") model.lattice.plot_brillouin_zone(decorate=False) bands.plot_kpath(point_labels=[r"$\Gamma$", "K", "M", r"$\Gamma$"])
grid = plt.GridSpec(3, 2, hspace=0.4) plt.figure(figsize=(6.7, 8)) for square, name in zip(grid, ["MoS2", "WS2", "MoSe2", "WSe2", "MoTe2", "WTe2"]): model = pb.Model(group6_tmd.monolayer_3band(name), pb.translational_symmetry()) solver = pb.solver.lapack(model) k_points = model.lattice.brillouin_zone() gamma = [0, 0] k = k_points[0] m = (k_points[0] + k_points[1]) / 2 plt.subplot(square, title=name) bands = solver.calc_bands(gamma, k, m, gamma) bands.plot(point_labels=[r"$\Gamma$", "K", "M", r"$\Gamma$"], lw=1.5)